Overview#
Introduction#
ClearMap is a toolbox for the analysis and registration of volumetric data from cleared tissues.
ClearMap has been designed to analyze terabyte-sized 3D datasets obtained via light sheet microscopy from iDISCO+ cleared tissue samples immunolabeled for proteins.
ClearMap has been written for mapping immediate early genes [Renier2016] as well as vasculature networks of whole mouse brains [Kirst2020]
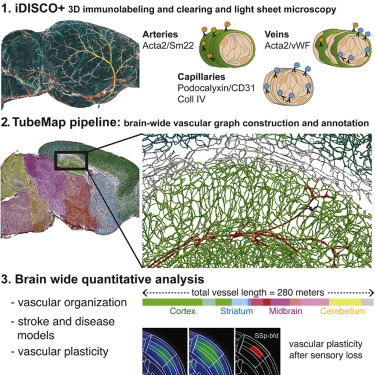
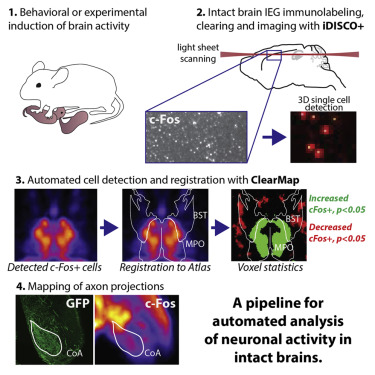
ClearMap tools may also be useful for data obtained with other types of microscopes, types of markers, clearing techniques, as well as other species, organs, or samples.
ClearMap is written in Python 3 and is designed to take advantage of parallel processing capabilities of modern workstations. We hope the open structure of the code will enable many new modules to be added to ClearMap to broaden the range of applications to different types of biological objects or structures.
Functionality and Pipelines#
ClearMap provides a larger set of high performance 3d image processing functions that are introduced here: Functionality.
These functions are combined together in expert functions under
Experts which are made use of in
pipelines found in Scripts to quantify the data sets.
The main tools are:
Usage#
An introduction of how to use ClearMap can be found here:
Documentation#
The full code documentation can be found here:
Source#
License#
GNU GENERAL PUBLIC LICENSE Version 3

Social Media#