cell_map#
This module contains the class to analyze (detect) individual cells, e.g. to analyze immediate early gene expression data from iDISCO+ cleared tissue [Renier2016].
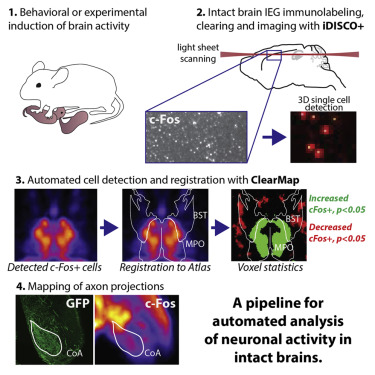
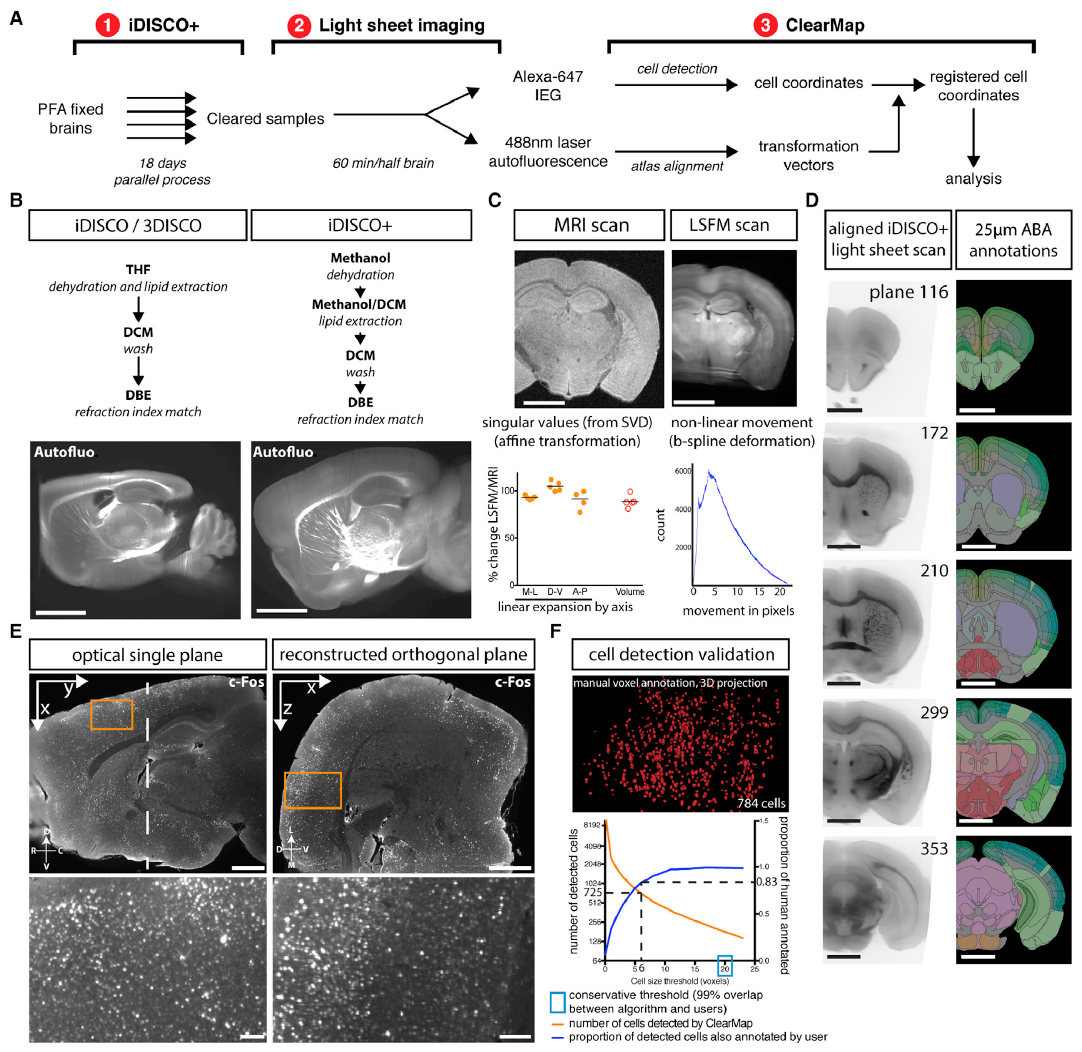
iDISCO+ and ClearMap: A Pipeline for Cell Detection, Registration, and Mapping in Intact Samples Using Light Sheet Microscopy.#
- class CellDetector(preprocessor=None)[source]#
Bases:
TabProcessor- export_as_csv()[source]#
Export the cell coordinates to csv
Deprecated since version 2.1: Use
atlas_align()and export_collapsed_stats instead.
- export_to_clearmap1_fmt()[source]#
ClearMap 1.0 export (will generate the files cells_ClearMap1_intensities, cells_ClearMap1_points_transformed, cells_ClearMap1_points necessaries to use the analysis script of ClearMap1. In ClearMap2 the ‘cells’ file contains already all this information) In order to align the coordinates when we have right and left hemispheres, if the orientation of the brain is left, will calculate the new coordinates for the Y axes, this change will not affect the orientation of the heatmaps, since these are generated from the ClearMap2 file ‘cells’
Deprecated since version 2.1: Use
atlas_align()and export_collapsed_stats instead.
- voxelize_unweighted(coordinates, voxelization_parameter)[source]#
Voxelize un weighted i.e. for cell counts
- Parameters:
coordinates – str, array or Source Source of point of nxd coordinates.
voxelization_parameter – dict
Returns
- voxelize_weighted(coordinates, source, voxelization_parameter)[source]#
Voxelize weighted i.e. for cell intensities
- Parameters:
coordinates – np.array
source – Source.Source
voxelization_parameter – dict
Returns
- property detected#
- property df_path#
