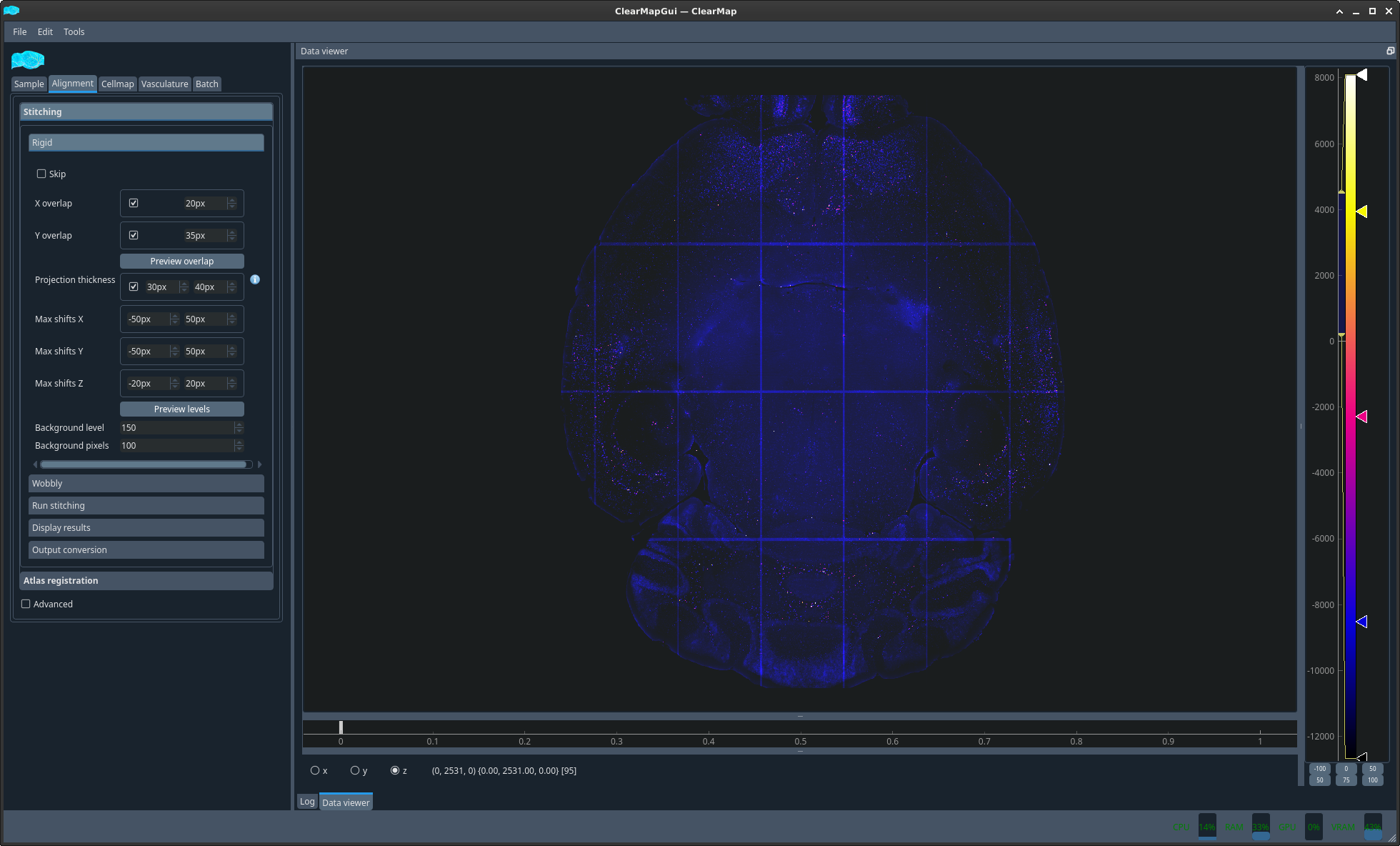
Atlas registration → Atlas settings tab#
To segment your data, i.e. assign a brain region to detected cells, you will need to align an atlas to your sample. This is done by aligning a reference image onto which an atlas was drawn and which was acquired with the same modality as your sample to a dedicated channel of your brain which contains only background information and no specific data. This channel is the autofluorescence channel. Because this channel is not acquired concomitantly to your data channels in light sheet microscopy, you will also need to align your autofluorescence channel to your data channel. Since both images come from the same sample, this second step is usually much simpler and only requires rigid registration, which is a combination of rotation, translation, scaling and shearing. On the contrary, the reference to autofluorescence alignment can be more challenging and uses a step of rigid registration followed by a step of elastic registration which involves local deformations of the sample.

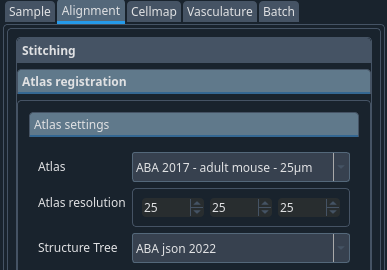
Atlas: ABA 2017 – adult mouse – 25 um
Atlas resolution: 25 25 25
Structure tree: ABA json 2022
Note
In future releases, these will become the defaults for the atlas so you won’t have to select them.
Tip
If you want to make these your defaults. You can do so by changing the value in the $HOME/.clearmap/default_alignment_params.cfg file in the relevant section.
Atlas registration → Run registration tab#
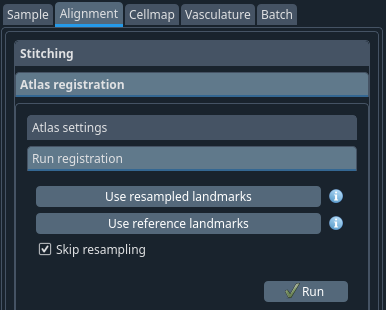
Usually. If the sample -> atlas space info parameters were defined correctly, it should suffice to click run. You can then verify the alignment in the next tab (see below).

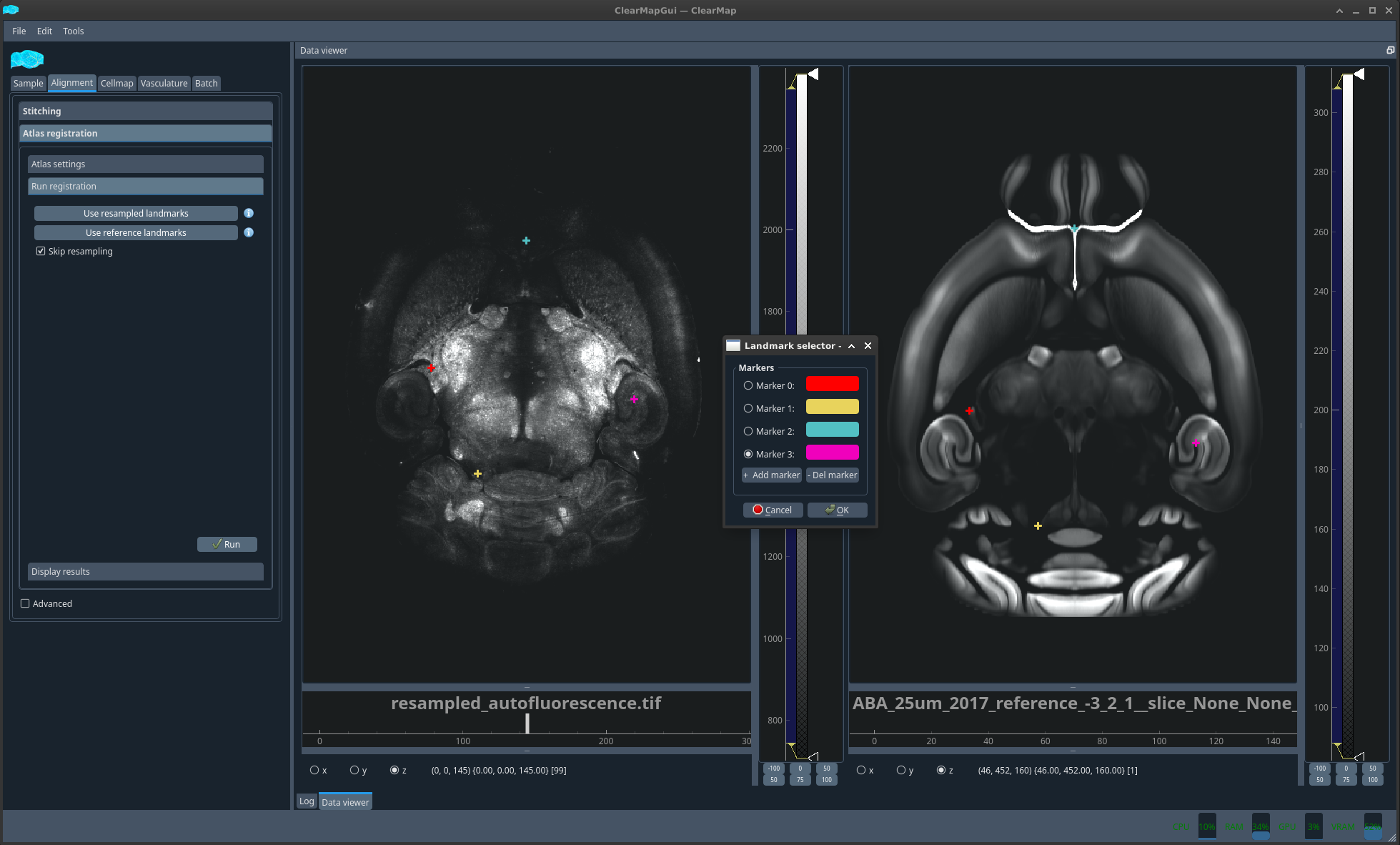
Should the alignment not be satisfactory, you could then select Use reference landmarks. A new dialog will appear together with the two images you are trying to align plotted side by side. You should click a feature on the left image that you can also easily pinpoint in the right image and the select add marker to create a new marker and repeat the operation. The number of markers required depends on the difficulty of alignment. For simple alignment mistakes, a handful of markers should suffice. However, this method was successfully used to align whole head MRI scans to LSM brain scans.
In the very unlikely event that the autofluorescence to raw data alignment is not correct, you could do the same with the Use resampled landmarks.

Atlas registration → Display results tab#
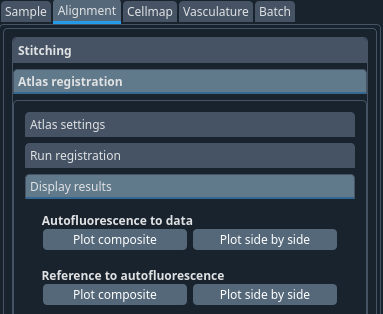
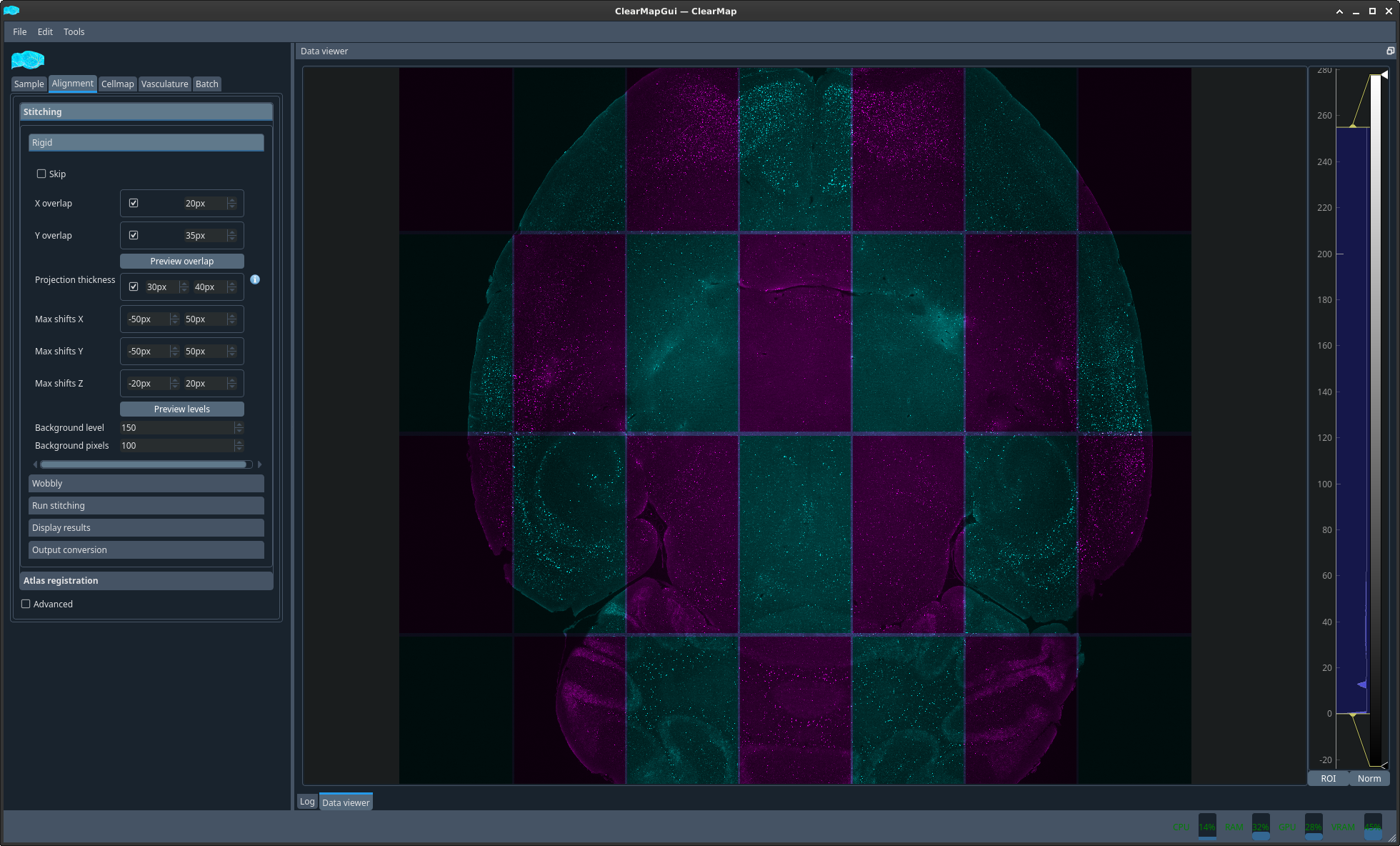
You can now display the result of the 2 different alignments discussed above in 2 different ways. Either as an overlay with 2 different colors or side by side with a matching crosshair.
Before proceeding with the analysis you should ensure that the alignments are correct, especially the more troublesome Reference to autofluorescence.

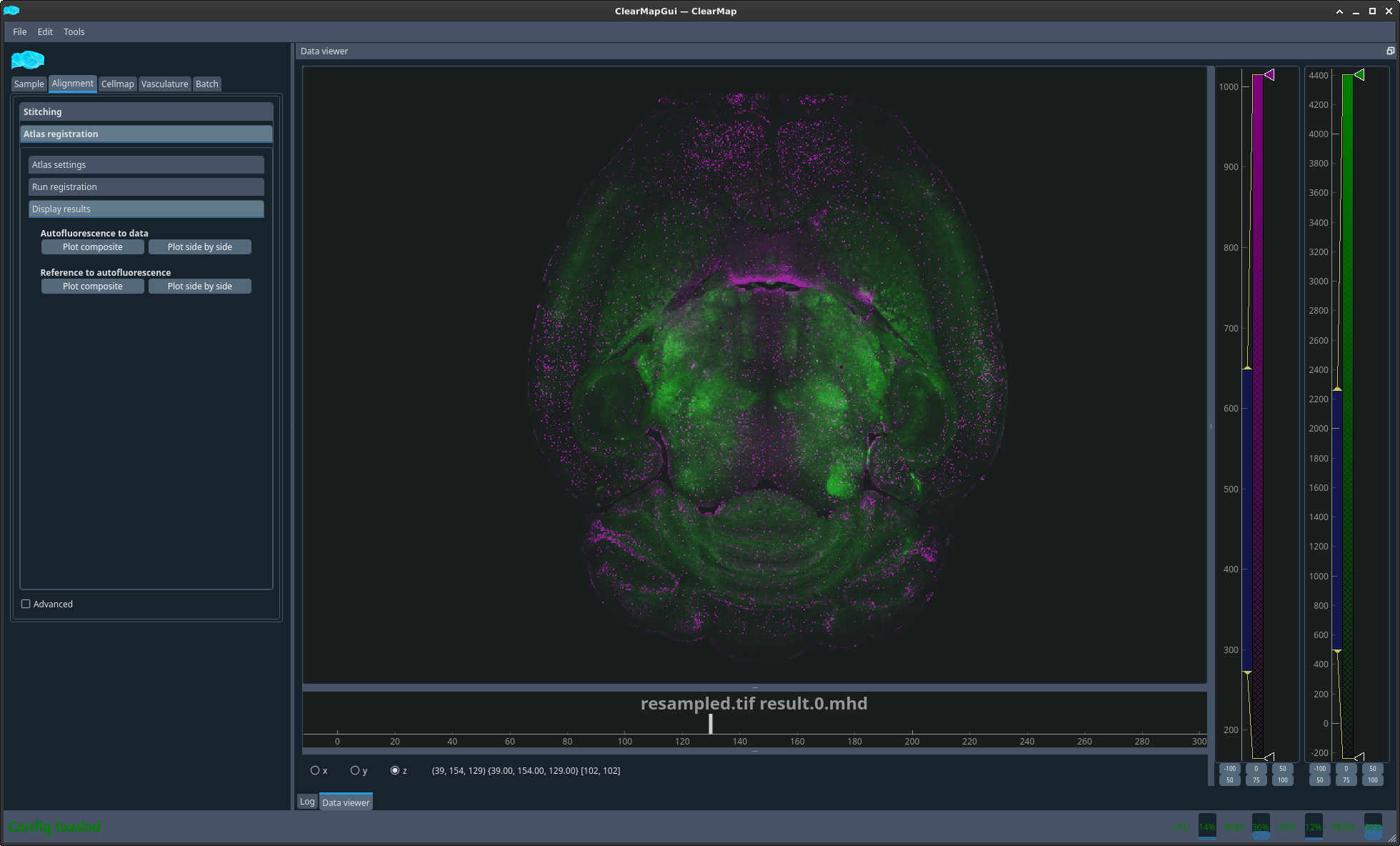
Figure 1 Plot auto to raw composite#
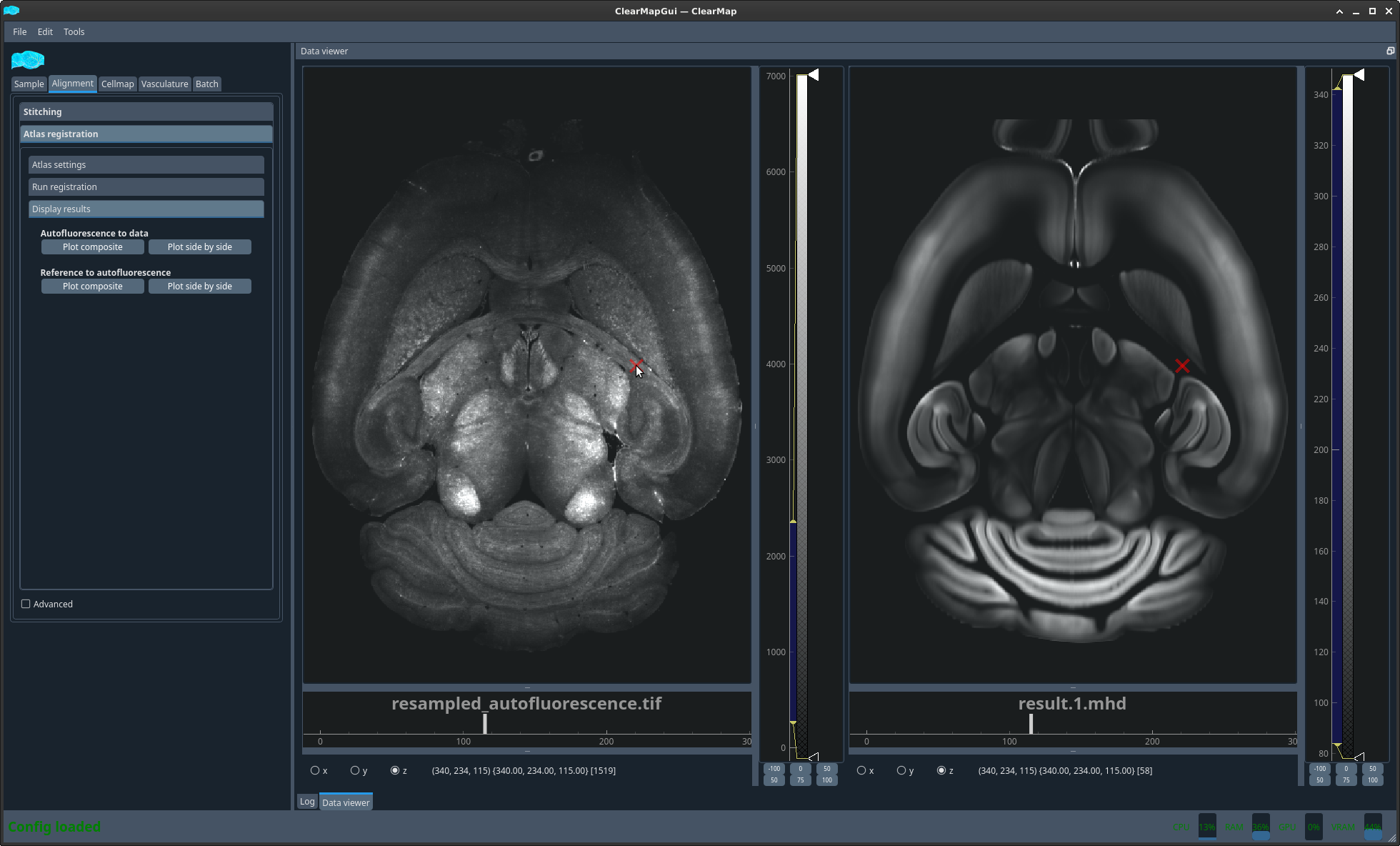
Figure 2 Plot ref to auto side by side#